Abstract
Introduction
Children with Down Syndrome (DS) frequently develop hematological malignancies including acute lymphoblastic leukemia (ALL), however, the genomic basis for the predisposition to DS-ALL remains unknown. DS-ALL is associated with increased rates of relapse, chemotherapy related toxicity and poor overall survival compared to non-DS patients. The P2RY8-CRLF2 (CRLF2r) gene fusion, the result of deletion of the pseudoautosomal region 1 (PAR1) on the X/Y chromosome (chr), has been identified in 60% of DS-ALL (+21) patients and 30% of iAMP21 (intrachromosomal amplification of chr21) ALL patients compared to only 5-16% of other B-ALL patients. The high mobility group nucleosome binding protein 1, encoded by HMGN1 on chr21 may play a role in DS-ALL leukemogenesis, due to its demethylase activity associated with transcriptional activation.
Methods
mRNA seq was performed on the blast cells of 76 ALL patients using the Universal Plus mRNA-seq with NuQuant kit. FusionCatcher, SOAPfuse and JAFFA were used to identify fusions and featureCounts used for mRNA expression of STAR (v2.7.3a) aligned BAM files. CRISPR/Cas9 gRNAs targeting P2RY8 intron 1 or CRLF2 5' UTR created the 320 KB (PAR1) deletion. Cas9/gRNAs were transduced into Jurkat cells ± HMGN1 overexpression, activated with doxycycline and stained with TSLPR (CRLF2/IL-7Rα dimer) for single cell sort and clonal expansion. Phosphorylated proteins were measured by intracellular flow cytometry and SYBR Green RQ-PCR was used for mRNA expression.
Results
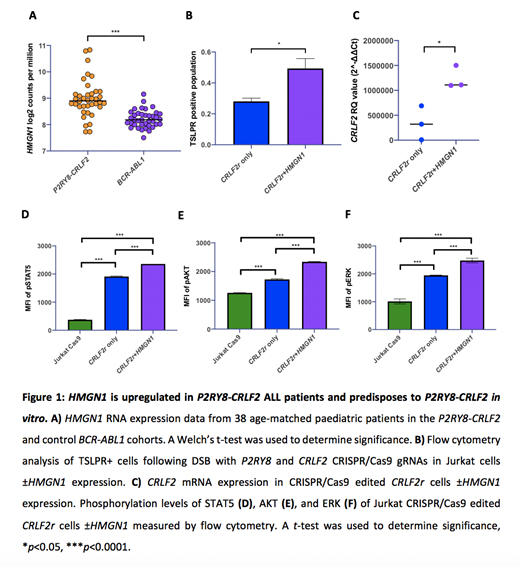
Significantly higher HMGN1 expression was identified in CRLF2r ALL patients, compared to the control (BCR-ABL1+ patients. Fig. 1A; p<0.0001). There was no significant difference in HMGN2 (p=0.7881) or JAK2 (p=0.1171) control genes between groups. Of the CRLF2r ALL patients, 26% harbored +21, and 5% iAMP21, strongly suggesting a relationship between chr21, HMGN1 and the CRLF2r fusion.
To further understand the role of HMGN1 in CRLF2r development, HMGN1 was overexpressed (1.5-fold; HMGN1H) in Jurkat Cas9 cells to represent a trisomy level of expression. gRNAs targeting P2RY8 and CRLF2 were activated in cells ± HMGN1H with undirected repair. TSLPR surface and CRLF2 mRNA expression were higher in CRLF2r + HMGN1H cells compared to CRLF2r only cells (Fig. 1B,C; p=0.034 and <0.001 respectively). This suggests higher HMGN1 expression significantly predisposed to PAR1 deletion and formation of CRLF2r fusion after a double stranded DNA break (DSB) versus repair where HMGN1 expression is normal. This is consistent with higher HMGN1 expression observed in patients with CRLF2r. There was also increased gene editing activity in CRLF2r + HMGN1H cells, identified through T7 gene editing assay, compared to CRLF2r only cells.
CRLF2r + HMGN1H cells also demonstrated significantly increased pSTAT5 (MFI: 2359 ± 1; Fig. 1D), pAKT (MFI: 2339 ± 6; Fig. 1E) and pERK (MFI: 2478 ± 48; Fig. 1F) compared to CRLF2r only cells (MFI pSTAT5: 1910 ± 10; pAKT: 1727 ± 14; pERK: 1946 ± 6; all p<0.001), consistent with the data observed in CRLF2r patients, and confirming cooperation between CRLF2 and HMGN1. CRLF2r + HMGN1H cells demonstrated reduced H3K27me3 (MFI: 1 ± 0.07) compared to CRLF2r only cells (MFI: 1.6 ± 0.03; p=0.003) and higher expression levels of STAT5 target genes (BCL2, CDKN1,MCL1and MYC).
Conclusion
The P2RY8-CRLF2 (CRLF2r) gene fusionis prevalent in DS, +21 and iAMP21 ALL patients. Here, we demonstrate that CRLF2r is associated with high expression of HMGN1 (chr21)in ALL patient cells . Using CRISPR/Cas9 in an in vitro model, we demonstrate that enforced high expression of HMGN1 alters DSB repair mechanism, favoring PAR1 deletion and the subsequent formation of the P2RY8-CRLF2 gene fusion, with attendant higher expression of STAT5 target genes. Understanding the role of HMGN1 in the disproportionate number of DS-ALL patients who are diagnosed with CRLF2r has the potential to lead to novel therapeutic interventions, in this high-risk group of patients where efficacious therapeutic options are currently poor.
Yeung: BMS: Honoraria, Research Funding; Amgen: Honoraria; Novartis: Honoraria, Research Funding; Pfizer: Honoraria. White: Novartis: Research Funding; BMS: Honoraria, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal